It is no secret that the scientific Python ecosystem is extensive, and even overwhelming for users at times. This is especially true for people coming from R, where the tidyverse has constructed an opinionated world in which there is just one tool needed to do any task. So what is it about the network of libraries in scientific Python that makes people want to use them even though it can be daunting? Spoiler: I think it has to do with extensibility.
Since I have been spending most of my time recently working on Python STAC libraries, I am also curious about how the approach in STAC borrows and differs from the more general scientific Python approach; in particular the practice of explicitly writing specifications before and during library development.
I recently had the opportunity to attend the SciPy conference in Austin, Texas, and Michael Droettboom’s keynote talk “Open Source Contributions through Time and Space” really resonated with me. He noted how improvements in packaging have made it easier and easier to distribute libraries. Furthermore, he discussed how more easily distributable libraries increase the likelihood that users will create a new library, rather than bloating existing libraries with new functionality.
As a maintainer, this is appealing because the new library can be maintained and released separately from the core library. The maintainers of the core library do not have to know that the extension exists. With separate authors and less emphasis on backward compatibility, these extension libraries can move more quickly than the core library can.
In addition to its benefits for maintainers, extensibility also benefits users as it is more likely that a user’s domain-specific algorithms will be packaged up by someone in the same field who has a solid understanding of how they work. It is also easier to move from user to contributor because it is easier to understand the codebase and feel ownership over it.
Given how straightforward and useful it is to create new modules in Python, it is unsurprising that the ecosystem has become a dense web of interdependent packages. In this reality, the challenge from a maintenance perspective becomes: How can we expose well-supported public APIs that other libraries can build off of?
In scientific Python, normally libraries are created and as they gain popularity the interface becomes the de facto standard. This is why libraries like Dask mimic the pandas and the NumPy APIs.
STAC takes the opposite approach by starting with a community-built specification for how to store and expose geospatial metadata.
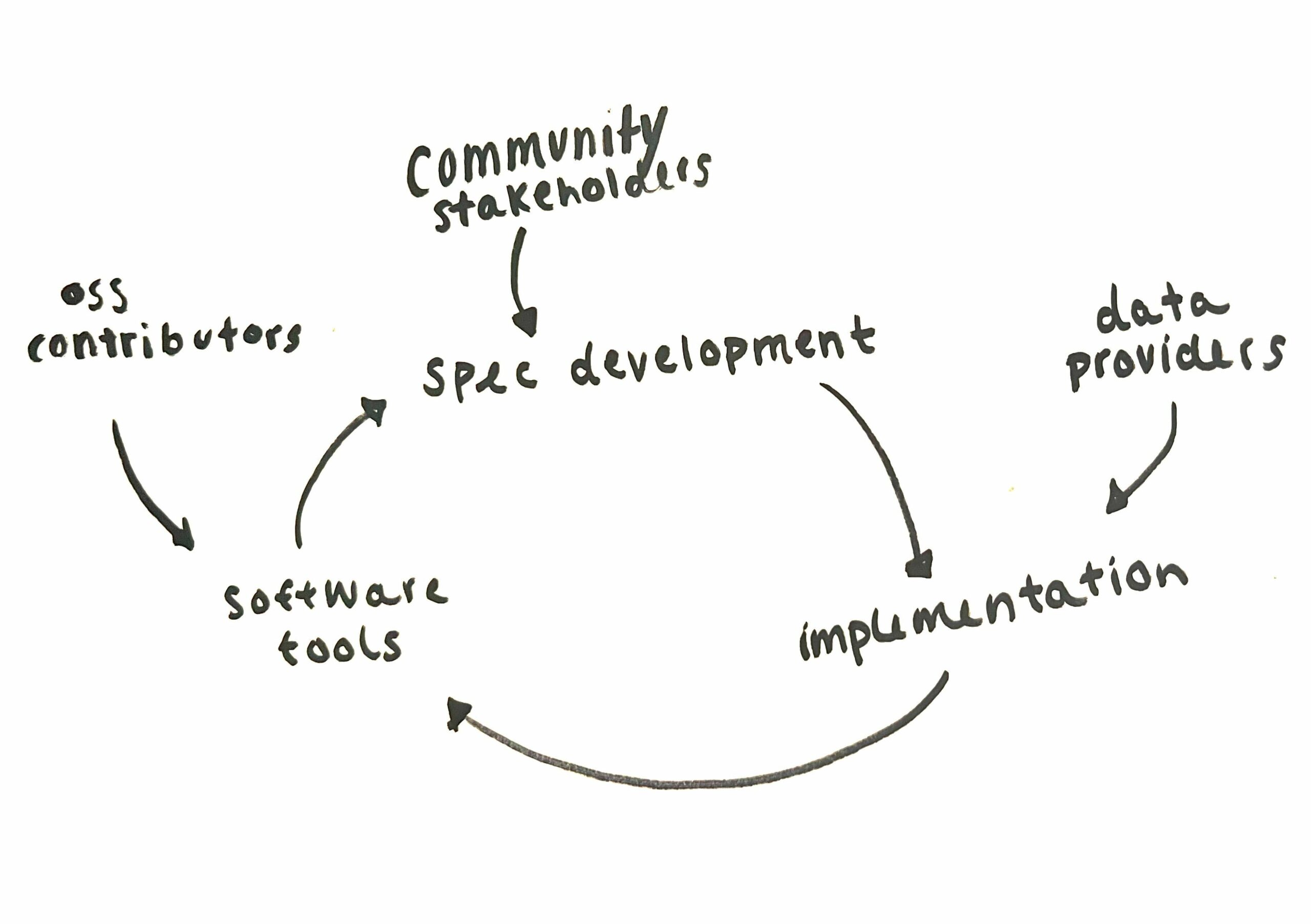
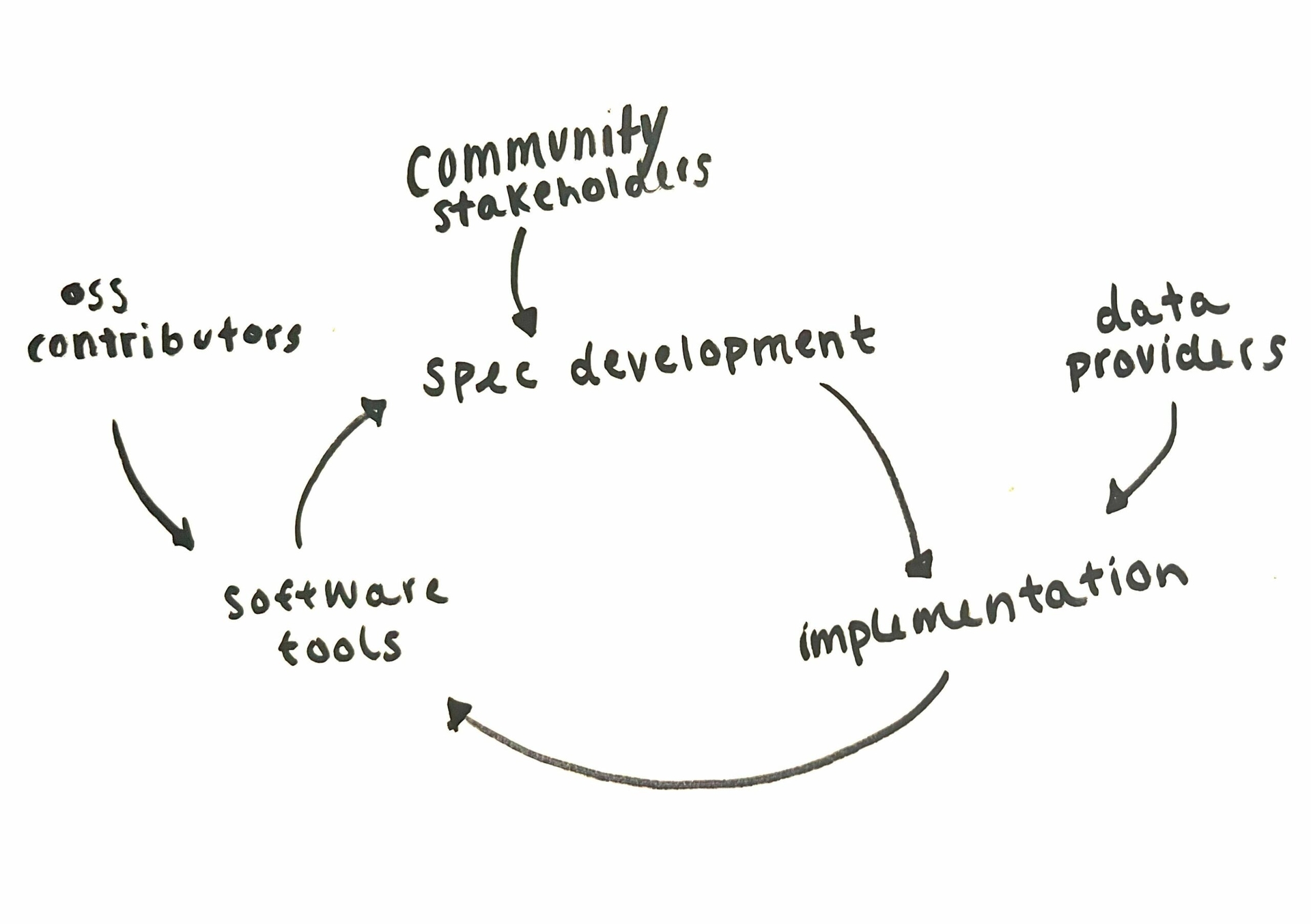
STAC is ruled by two core specifications: the STAC specification and the STAC API specification. Each of those specs has a mechanism for extending the spec to describe niche STAC metadata or the behavior of new endpoints on the STAC API. Open source developers take those specs and build tools to support them. Data curators who are tasked with setting up STAC catalogs implement the core, and then get to choose which extensions make sense for their data and users. Data curators can even create new extensions if necessary. Specification development, library development, and implementation can all happen at the same time and naturally inform each other.
Similar to scientific Python itself, part of what makes STAC appealing is the multitude of libraries that have sprung up to satisfy the requirements of the specs. On the data consumer side, there are core libraries for accessing STAC metadata from Python (PySTAC) and for accessing STAC APIs (PySTAC-Client). These libraries form the core of the python interface for STAC. Then there are other libraries that build off this core and let you create xarray objects out of STAC items (odc-stac, stackstac). This same functionality is actually implemented in two different ways in two different libraries. This can be seen as duplicated effort, but in another light it shows the power of modularity. Since the functionality that the libraries need to provide is so well-described, we can fairly easily swap one approach out for another. Either way we start with STAC metadata and end up with an xarray data structure.
STAC’s explicit modularization in both the tools and the spec enables parallel streams of tool development, spec development, and implementation and encourages everyone to participate in all parts of the development process. You can read more from Matt Hanson about the history of STAC and start getting excited for the next segment in that series – coming out soon!
You can find a description of the keynote talk here, and you can also view the full recording now on YouTube. Thanks to Numfocus for putting on a great SciPy! I can’t wait for next year!